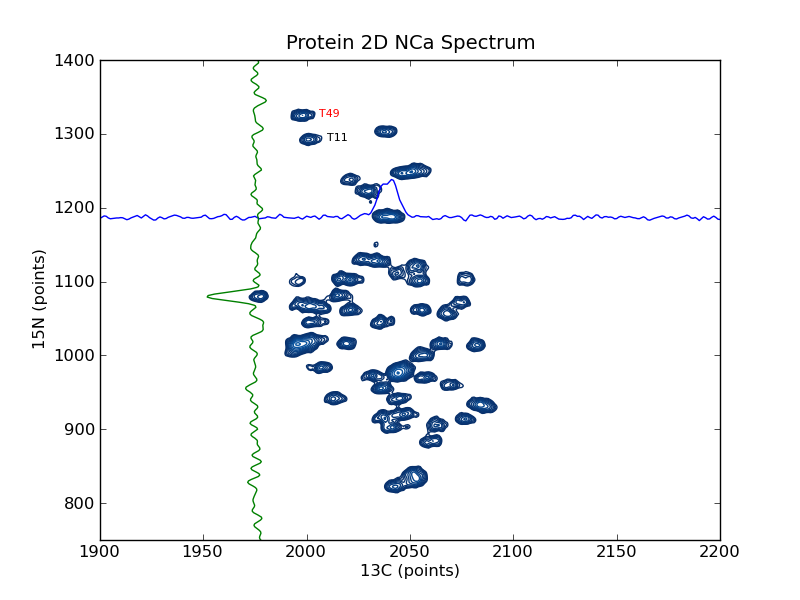
plotting example: plot_2d_spectrum_pts¶
This example shows how to use nmrglue and matplotlib to create figures for examining data or publication. In this example a contour plot of the spectrum from a 2D NMRPipe file is created. Slices are added in the 15N and 13C dimension as well as sample peak labels. plotting example: plot_2d_spectrum is similar to this example but plotted on a ppm scale.
#! /usr/bin/env python
# Create contour plots of a 2D NMRPipe spectrum
import nmrglue as ng
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.cm
# plot parameters
cmap = matplotlib.cm.Blues_r # contour map (colors to use for contours)
contour_start = 30000 # contour level start value
contour_num = 20 # number of contour levels
contour_factor = 1.20 # scaling factor between contour levels
# calculate contour levels
cl = [contour_start*contour_factor**x for x in xrange(contour_num)]
# read in the data from a NMRPipe file
dic,data = ng.pipe.read("../../common_data/2d_pipe/test.ft2")
# create the figure
fig = plt.figure()
ax = fig.add_subplot(111)
# plot the contours
ax.contour(data,cl,cmap=cmap,extent=(0,data.shape[1]-1,0,data.shape[0]-1))
# add some labels
ax.text(2006,1322,"T49",size=8,color='r')
ax.text(2010,1290,"T11",size=8,color='k')
# plot slices in each direction
xslice = data[1187,:]
ax.plot(xrange(data.shape[1]),xslice/3.e3+1187)
yslice = data[:,1976]
ax.plot(-yslice/3.e3+1976,xrange(data.shape[0]))
# decorate the axes
ax.set_ylabel("15N (points)")
ax.set_xlabel("13C (points)")
ax.set_title("Protein 2D NCa Spectrum")
ax.set_xlim(1900,2200)
ax.set_ylim(750,1400)
# save the figure
fig.savefig("spectrum_pts.png") #change to .pdf, .ps, etc for different formats
Result: