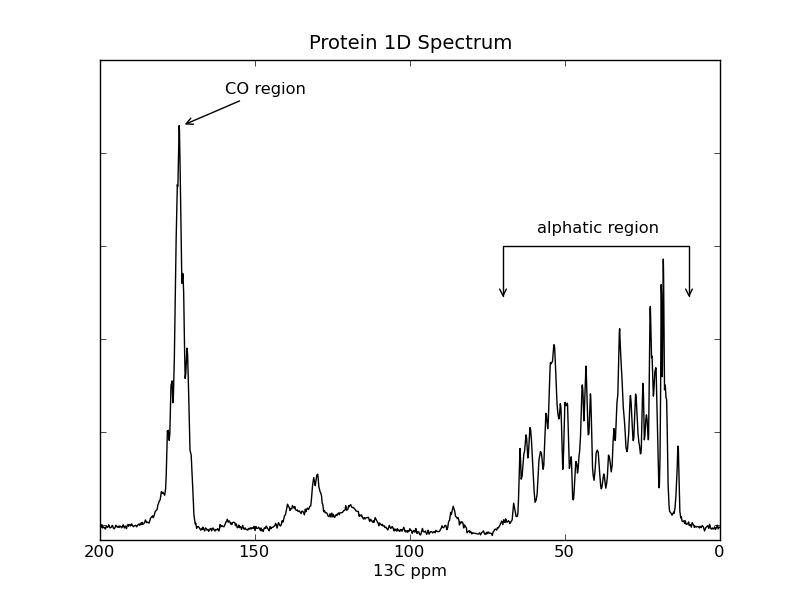
plotting example: plot_1d_freq¶
This example shows how to use nmrglue and matplotlib to create figures for examining data or publication. In this example the spectrum from a 1D NMRPipe file is plotted.
#! /usr/bin/env python
# Create a 1D plot of NMRPipe data
import nmrglue as ng
import matplotlib.pyplot as plt
import numpy as np
# read in the data from a NMRPipe file
dic,data = ng.pipe.read("../../common_data/1d_pipe/test.ft")
# create a unit conversion object for the axis
uc = ng.pipe.make_uc(dic,data)
# plot the spectrum
fig = plt.figure()
ax = fig.add_subplot(111)
ax.plot(uc.ppm_scale(),data,'k-')
# annotate the figure
ax.annotate('CO region',xy=(173,2.15e6),xycoords='data',
xytext=(30,20),textcoords='offset points',
arrowprops=dict(arrowstyle="->") )
ax.text(59,1.55e6,"alphatic region")
ax.annotate('',xy=(70,1.2e6),xycoords='data',
xytext=(10,1.2e6),textcoords='data',
arrowprops=dict(arrowstyle="<->",
connectionstyle="bar",
ec="k",
shrinkA=5,shrinkB=5,))
# decorate axes
ax.set_yticklabels([])
ax.set_title("Protein 1D Spectrum")
ax.set_xlabel("13C ppm")
ax.set_xlim(200,0)
ax.set_ylim(-80000,2500000)
# save the figure
fig.savefig("spectrum.png") # change this to .pdf, .ps, etc
Results:
[figure]