plotting example: plot_2d_boxes¶
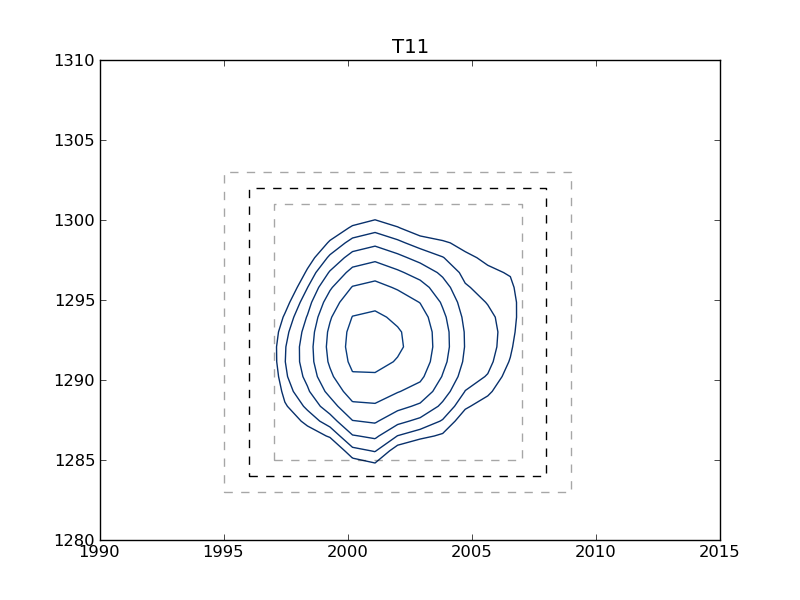
This example shows how to use nmrglue and matplotlib to create figures for examining data or publication. In this example the box limits used in integration example: integrate_2d are graphically examined. A contour plot of each peak is plotted with the box limits indicated by the dark dashed line. To check peak assignments see plotting example: plot_2d_assignments.
#! /usr/bin/env python
# Create contour plots of each peak defined in limits.in file
import nmrglue as ng
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.cm
# plot parameters
xpad = 5 # padding around peak box on x-axis
ypad = 5 # padding around peak box on y-axis
cmap = matplotlib.cm.Blues_r # contour map (colors to use for contours)
contour_start = 30000 # contour level start value
contour_num = 20 # number of contour levels
contour_factor = 1.20 # scaling factor between contour levels
# calculate contour levels
cl = [contour_start*contour_factor**x for x in xrange(contour_num)]
# read in the data from a NMRPipe file
dic,data = ng.pipe.read("../../common_data/2d_pipe/test.ft2")
# read in the integration limits
peak_list = np.recfromtxt("limits.in")
# loop over the peaks
for name,x0,y0,x1,y1 in peak_list:
if x0>x1:
x0,x1 = x1,x0
if y0>y1:
y0,y1 = y1,y0
# slice the data around the peak
slice = data[y0-ypad:y1+1+ypad,x0-xpad:x1+1+xpad]
# create the figure
fig = plt.figure()
ax = fig.add_subplot(111)
# plot the contours
print "Plotting:",name
etup = (x0-xpad+1,x1+xpad-1,y0-ypad+1,y1+ypad-1)
ax.contour(slice,cl,cmap=cmap,extent=etup)
# draw a box around the peak
ax.plot([x0,x1,x1,x0,x0],[y0,y0,y1,y1,y0],'k--')
# draw light boxes at +/- one point
ax.plot([x0-1,x1+1,x1+1,x0-1,x0-1],[y0-1,y0-1,y1+1,y1+1,y0-1],
'k--',alpha=0.35)
ax.plot([x0+1,x1-1,x1-1,x0+1,x0+1],[y0+1,y0+1,y1-1,y1-1,y0+1],
'k--',alpha=0.35)
# set the title
ax.set_title(name)
# save the figure
fig.savefig(name+".png")
del(fig)
#Peak X0 Y0 X1 Y1
# Peak defines 15N resonance in 2D NCO spectra.
# Limits are in term of points from 0 to length-1.
# These can determined from nmrDraw by subtracting 1 from the X and Y
# values reported.
#Peak X0 Y0 X1 Y1
T49 1992 1334 2003 1316
T11 1996 1302 2008 1284
# comments can appear anywhere in this file just start the line with #
G14 2032 1314 2044 1293
E15 2077 1025 2087 1004
W43 2008 952 2019 933
Sample Figure
[T11.png]